 During my research regarding Covid-19, I came across this study. While I found the information compelling, I honestly did not know how to write an article about it, given the amount of data it sourced. Rather than give up, I decided to simply write one of the major conclusions in the title of this piece. I will repeat it here:
During my research regarding Covid-19, I came across this study. While I found the information compelling, I honestly did not know how to write an article about it, given the amount of data it sourced. Rather than give up, I decided to simply write one of the major conclusions in the title of this piece. I will repeat it here:
Ashkenazi Jews and Amish Are Least Susceptible to Covid-19
The study is entitled:
New Insights into Genetic Susceptibility Of COVID-19: An ACE2 And TMPRSS2 Polymorphism Analysis published by BMC Medicine.
Hou, Y., Zhao, J., Martin, W. et al. new insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med 18, 216 (2020).
I do not claim to have any medical knowledge. I am not a doctor, epidemiologist, nor a researcher. The key takeaway for me is that the immune receptor called ACE2 is not susceptible to any disruption by the Covid-19 virus in Ashkenazi Jews.
Anti-Semites have used parts of this study to develop a conspiracy theory where they are convinced that Covid-19 is a plot by Jews to dominate the world. Anti-Semites have used this canard for almost everything under the sun, from Jews controlling banking, Wall Street and the so-called “entertainment industry” (Motion pictures and recording), for just a few examples.
The following are unedited excerpts from the actual study and all the sources for those who may be capable of understanding them. Again, the bottom line is: the immune receptor called ACE2 is not susceptible to any disruption by the Covid-19 virus in Ashkenazi Jews.
You can draw your own conclusions, as for me, it means that cases of Covid-19 would be mild, not lasting long, and would develop natural immunity and thus herd immunity across the Ashkenazi population. You can see the graphic results of the study below in Fig. 1b.
ACE2 polymorphism analysis across different populations. Here, we investigated genetic susceptibility to COVID19 by examining DNA polymorphisms in ACE2 (OMIM 300335) and TMPRSS2 (OMIM 602060) genes. We assembled a total of 437 non-synonymous singlenucleotide variants (SNVs) in the protein-coding regions of ACE2 and TMPRSS2 (Fig. 1a) from three databases: (i) Genome Aggregation Database (gnomAD v3: gnomad.broadinstitute.org, covering 9 geographical areas), (ii) Exome Sequencing Project (ESP: evs.gs.washington. edu/EVS/), and (iii) 1000 Genomes Project (1KGP, www. internationalgenome.org). We used ANNOVAR [9] to annotate all non-synonymous variants. By applying Polyphen2 and CADD (Combined Annotation Dependent Depletion) scores, we identified 63 potentially deleterious variants in ACE2 (61 in gnomAD) and 68 deleterious variants in TMPRSS2 (63 in gnomAD). We found that the distribution of deleterious variants in ACE2 differs among 9 populations in gnomAD (v3). Specifically, 39% (24/61) and 54% (33/61) of deleterious variants in ACE2 occur in African/African-American (AFR) and Non-Finnish European (EUR) populations, respectively (Fig. 1b). Prevalence of deleterious variants among Latino/Admixed American (AMR), East Asian (EAS), Finnish (FIN), and South Asian (SAS) populations is 2–10%, while Amish (AMI) and
Ashkenazi Jewish (ASJ) populations do not appear to carry such variants in ACE2 coding regions
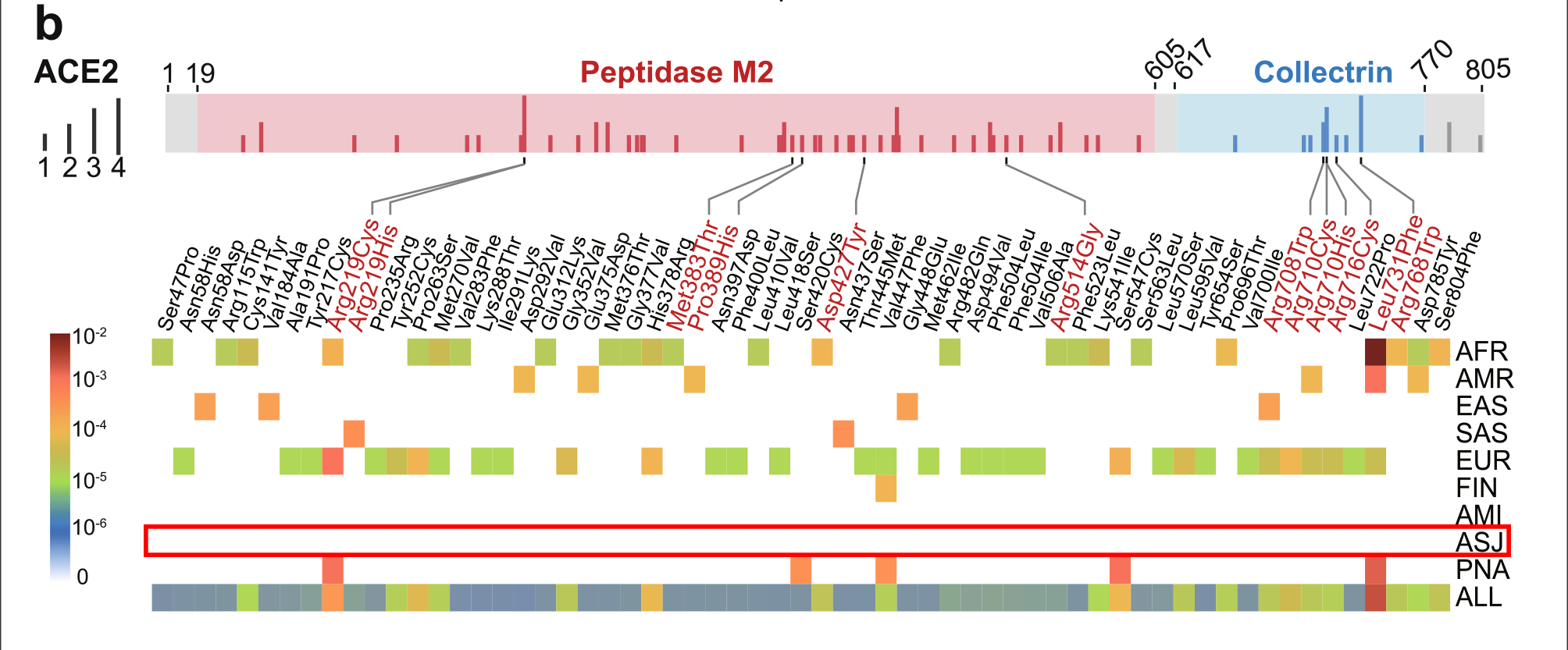
Fig. 1: The coding-region variants in ACE2 and TMPRSS2 from ~ 81,000 human genomes across 8 populations. a Coding-region variants in the genes encoding angiotensin-converting enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2) across three human genome databases: (i) Genome Aggregation Database (gnomAD v3), (ii) Exome Sequencing Project (ESP), and (iii) 1000 Genomes Project (1KGP). SARS-CoV-2 utilizes the host cell factors angiotensin-converting enzyme 2 (ACE2) for entry into cells and the host transmembrane serine protease TMPRSS2 for SARS-CoV-2 spike (S) protein priming, offering potential pathway for therapeutic development in treatment of COVID-19. b Distribution of 61 deleterious variants in the ACE2 coding region identified in gnomAD (v3). Polyphen2 > 0.96 and CADD scores > 20 as cutoff identify putative deleterious variants. The upper panel using 3 colors shows the functional domains of ACE2, and the height of the vertical line represents the number of populations that carry this variant. The lower heatmap shows the allele frequencies (color key) of a variant across different populations. c Distributions of 63 putative deleterious variants in the TMPRSS2 coding region using the same approach of b. AFR, African/African-American; AMI, Amish; AMR, Latino/Admixed American; ASJ, Ashkenazi Jewish; EAS, East Asian; FIN, Finnish; EUR, Non-Finnish European; SAS, South Asian; PNA, population not assigned.
Conclusions
This comprehensive comparative genetic analysis of approximately 81,000 human genomes suggested possible associations of ACE2 and TMPRSS2 DNA polymorphisms with COVID-19 susceptibility, severity, and clinical outcomes. We found that ACE2 polymorphisms were more likely to be associated with cardiovascular and pulmonary conditions by altering the angiotensinogen-ACE2 interactions, such as p.Arg514Gly in the African/African-American population. Unique but prevalent polymorphisms in TMPRSS2, including p.Val160Met (rs12329760), may provide potential explanations for differential genetic susceptibility to COVID19 as well as for risk factors, including cancer and the high-risk group of male patients. We highlighted that polymorphisms in ACE2 or TMPRSS2 could guide personalized treatments (i.e., hydroxychloroquine and camostat) for COVID-19. In summary, this study suggested that ACE2 or TMPRSS2 DNA polymorphisms were likely associated with genetic susceptibility to COVID-19, which calls for a human genetics initiative for fighting the COVID-19 pandemic.
Availability of data and materials All population genetic data used in this study are free and available at three databases: (i) Genome Aggregation Database (gnomAD v3: gnomad. broadinstitute.org, covering 9 geographical areas), (ii) Exome Sequencing Project (ESP: evs.gs.washington.edu/EVS/), and (iii) 1000 Genomes Project (1KGP, www.internationalgenome.org).
Yuan Hou1, Junfei Zhao2, William Martin1, Asha Kallianpur1,3, Mina K. Chung3,4,5, Lara Jehi5, Nima Sharifi3,5, Serpil Erzurum3,5, Charis Eng1,3,6,7 and Feixiong Cheng1,3,7*
- Dong Y, Mo X, Hu Y, Qi X, Jiang F, Jiang Z, Tong S. Epidemiology of COVID-19 among children in China. Pediatrics. 2020;8(6):2118–20.
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91.
- Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–80.
- Stopsack KH, Mucci LA, Antonarakis ES, Nelson PS, Kantoff PW. TMPRSS2 and COVID-19: serendipity or opportunity for intervention? Cancer Discov. 2020; 10(6):779–82.
- Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020. https://doi.org/10.1007/s11684-020-0754-0.
- Guo T, Fan Y, Chen M, Wu X, Zhang L, He T, Wang H, Wan J, Wang X, Lu Z. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19). JAMA Cardiol. 2020. https://doi.org/10.1001/jamacardio.2020.1017.
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38(16):e164.
- Li W, Zhang C, Sui J, Kuhn JH, Moore MJ, Luo S, Wong SK, Huang IC, Xu K, Vasilieva N, et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24(8):1634–43.
- Kuster GM, Pfister O, Burkard T, Zhou Q, Twerenbold R, Haaf P, Widmer AF, Osswald S. SARS-CoV2: should inhibitors of the renin-angiotensin system be withdrawn in patients with COVID-19? Eur Heart J. 2020;41(19):1801–3.
- Heurich A, Hofmann-Winkler H, Gierer S, Liepold T, Jahn O, Pohlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014;88(2):1293–307.
- Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015.
348(6235):648–60.
- Zheng Z, Huang D, Wang J, Zhao K, Zhou Y, Guo Z, Zhai S, Xu H, Cui H, Yao H, et al. QTLbase: an integrative resource for quantitative trait loci across multiple human molecular phenotypes. Nucleic Acids Res. 2020;48(D1): D983–91.
- Shirato K, Kawase M, Matsuyama S. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology. 2018; 517:9–15.
- Yu J, Ouyang W, Chua MLK, Xie C. SARS-CoV-2 transmission in patients with cancer at a tertiary care hospital in Wuhan. China JAMA Oncol. 2020. https://doi.org/10.1001/jamaoncol.2020.0980.
- Schuler A, Habermann C, Plosa J, et al. Age-related expression of SARS-CoV-2 primining protease TMPRSS2 in the developing lung. 2020. https://doi. org/10.1101/2020.05.22.111187 bioRxiv preprint doi: https://doi.org/10.1101/ 2020.05.22.111187.
- Mostafavi H, Berisa T, Day FR, Perry JRB, Przeworski M, Pickrell JK. Identifying genetic variants that affect viability in large cohorts. PLoS Biol. 2017;15(9): e2002458.
- Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020; 6:14.
- Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020. https://doi.org/10.1001/jama.2020.6019.
- Savarino A, Di Trani L, Donatelli I, Cauda R, Cassone A. New insights into the antiviral effects of chloroquine. Lancet Infect Dis. 2006;6(2):67–9.
- Rosenberg ES, Dufort EM, Udo T, Wilberschied LA, Kumar J, Tesoriero J, Weinberg P, Kirkwood J, Muse A, DeHovitz J, et al. Association of treatment with hydroxychloroquine or azithromycin with in-hospital mortality in patients with COVID-19 in New York state. JAMA. 2020. https://doi.org/10. 1001/jama.2020.8630.
- Walls AC, Tortorici MA, Snijder J, Xiong X, Bosch BJ, Rey FA, Veesler D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci U S A. 2017;114(42):11157–62.
- Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. 2011;85(2): 873–82.
- Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci U S A. 2005;102(33):11876–81.
- Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;
367(6485):1444–8.
- Initiative C-HG. The COVID-19 host genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur J Hum Genet. 2020;28(6):715–8.
- Group TSC-G. Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med. 2020. https://doi.org/10.1056/NEJMoa2020283.
This work was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health (NIH) under Award Number R00HL138272 and the National Institute of Aging under Award Number R01AG066707 to F.C. This work was supported, in part, by the VeloSano Pilot Program (Cleveland Clinic Taussig Cancer Institute).
Author information
Affiliations
- Genomic Medicine Institute, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, 44195, USA
Yuan Hou, William Martin, Asha Kallianpur, Charis Eng & Feixiong Cheng
- Department of Systems Biology and Department of Biomedical Informatics, Herbert Irving Comprehensive Center, Columbia University, New York, NY, 10032, USA
Junfei Zhao
- Department of Molecular Medicine, Cleveland Clinic Lerner College of Medicine, Case Western Reserve University, Cleveland, OH, 44195, USA
Asha Kallianpur, Mina K. Chung, Nima Sharifi, Serpil Erzurum, Charis Eng & Feixiong Cheng
- Department of Cardiovascular Medicine, Heart, Vascular and Thoracic Institute, Cleveland Clinic, Cleveland, OH, 44195, USA
Mina K. Chung
- Lerner Research Institute, Cleveland Clinic, Cleveland, OH, 44195, USA
Mina K. Chung, Lara Jehi, Nima Sharifi & Serpil Erzurum
- Department of Genetics and Genome Sciences, School of Medicine, Case Western Reserve University, Cleveland, OH, 44106, USA
Charis Eng
- Case Comprehensive Cancer Center, School of Medicine, Case Western Reserve University, Cleveland, OH, 44106, USA
Charis Eng & Feixiong Cheng
https://doi.org/10.1186/s12916-020-01673-z
Creative Commons
This is an open access article distributed under the terms of the Creative Commons CC BY license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About BMC
BMC has an evolving portfolio of some 300 peer-reviewed journals, sharing discoveries from research communities in science, technology, engineering and medicine. In 1999 we made high quality research open to everyone who needed to access it – and in making the open access model sustainable, we changed the world of academic publishing.
We are committed to continual innovation in research publishing to better support the needs of our communities, ensuring the integrity of the research we publish and championing the benefits of open research for all.
Our leading research journals include selective titles such as BMC Biology, BMC Medicine, Genome Biology and Genome Medicine, academic journals such as Journal of Hematology & Oncology, Malaria Journal and Microbiome, and the BMC series, 65 inclusive journals focused on the needs of individual research communities. We also partner with leading institutions and societies to publish journals on their behalf.
BMC is part of Springer Nature, giving us greater opportunities to help authors everywhere make more connections with research communities across the world.


disclaimer: I did not read through this article, because I don’t understand the scientific terms. I am not disqualifying this, just adding my own opinion of why Jews and Amish likely have the lower cases of covid. This is because these 2 groups are least likely to test for covid when sick, so there are less documented cases – emphasize documented
As a medical doctor who drove through Amish country during the height of the pandemic, I can tell you that our resistance comes not from mutations to the ACE receptor, but due to the fact that the Amish and the Frum communities are the youngest demographic in the country, and also because both communities ignored the quarantine and masking mandates (the Amish more so than us), so were already exposed.
I can’t even count the number of sources in this article. Are you amongst them? Your sourcing a drive through Amish country? You have to be kidding. You are Dr. Moshe who? Sorry doc, your opinion is NOT “due to the [your] fact”. If you are truly a doctor, kindly post your practice name so I can avoid it at all costs.
These 2 demographics were least likely to take the covid vax. Period.
Anyone have a count of how many ashkenazi funerals we had??????????? Not to mention the amount if people that were deathly sick ????
Just here in Lakewood we are now in the period of yahrzeits almost daily from civic deaths
The reason, by and large that the frum community and the Amish have had a greater success with skirting Covid is because they both don’t have televisions. (and we also have a very healthy dose of skepticism)
Fear drives illness.
I did some research. Not all Yidden who go by Ashkenazi are truly genetically so. A DNA test can show genetic discrepancies. Mine shows somewhere we have Sephardi mutations. That makes sense because my great grandparents named Epstein were descendants of the Torah Temimah who was Sephardic.
Seems a bit too coincidental to my liking that the 2 most insular communities are the the most resistant. While certainly the virus, because is manmade can certainly have been engineered to target certain demographics… and anecdotal evidence does in fact support this as certain communities as the asian/black/latin were hit harder than Caucasians. But considering that ashkenazi jews and the amish are of very different backgrounds, it doesn’t seem probable. But then again… it’s not the first thing the chinese botched up on programming correctly… they aren’t great at their own creations.. they do much better when stealing American designs and copying them.
Weren’t they saying a short while ago that those Ashkenazi zip codes had the highest levels of covid? A bit confused.